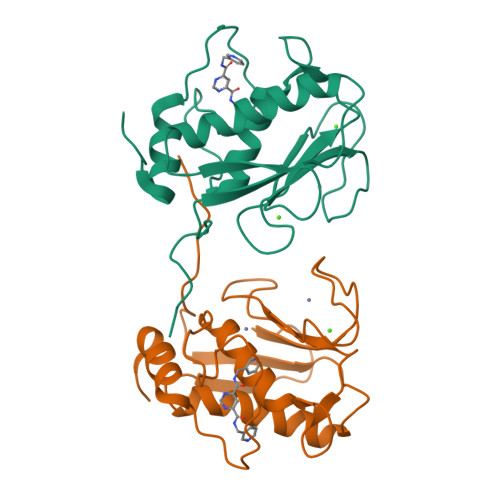
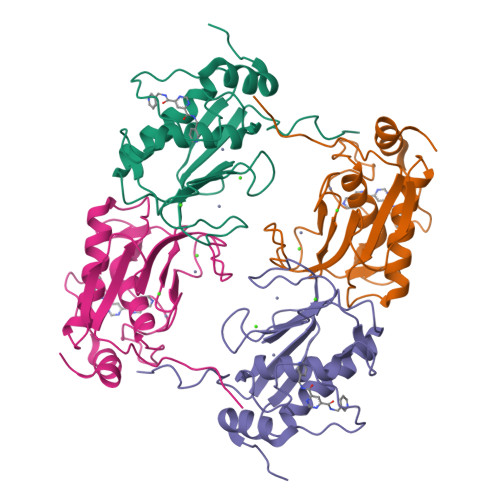
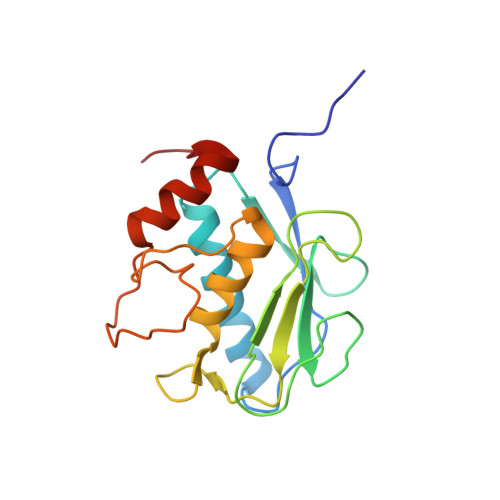
Structural basis for the highly selective inhibition of MMP-13.
Engel, C.K., Pirard, B., Schimanski, S., Kirsch, R., Habermann, J., Klingler, O., Schlotte, V., Weithmann, K.U., Wendt, K.U.(2005) Chem Biol 12: 181-189
- PubMed: 15734640
- DOI: https://doi.org/10.1016/j.chembiol.2005.01.008
- Primary Citation of Related Structures:
1XUD, 1XUR - PubMed Abstract:
The paradigm for matrix metalloprotease inhibition combines active site tailoring and catalytic zinc ligation. But, selectivity has been difficult. Now, Engel et al. present novel compounds, completely selective for MMP-13, with a unique binding mode.
Organizational Affiliation:
Incyte Corporation, Experimental Station, Route 141 and Henry Clay Road, Wilmington, Delaware 19880, USA